Page Not Found
Page not found. Your pixels are in another canvas. Read more
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas. Read more
About me Read more
This is a page not in th emain menu Read more
Published:
Useful resource for every coding enthusiast
A compilation of the most used commands while coding in Vim Read more
A compilation of the most used commands in bash Read more
Online summer school on machine learning and data sciences that we organized. Read more
Some of the common resources we use in computational pathology Read more
Some useful CS topics that are not taught as part of the university curriculum. The standard CS curriculum is missing some of these critical topics about the computing ecosystem that could make students lives significantly easier. Read more
Published in 2016 International Symposium on Intelligent Signal Processing and Communication Systems (ISPACS), 2016
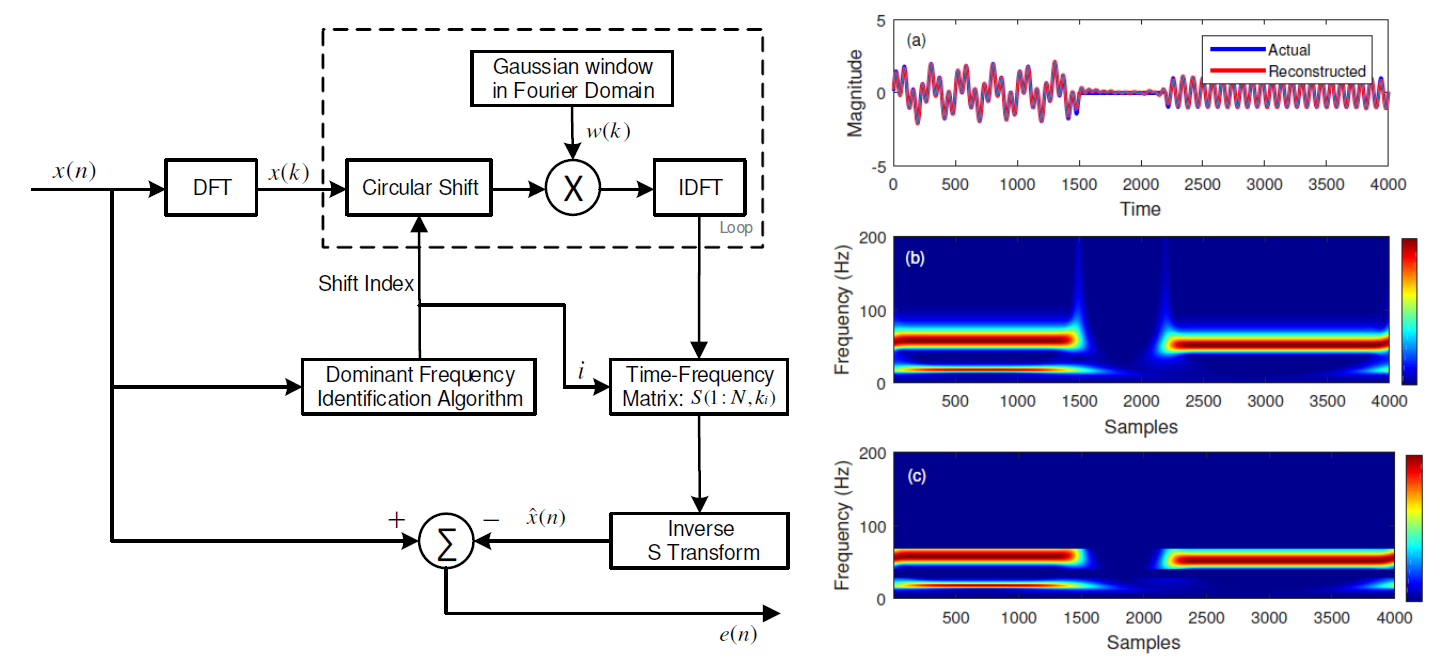
This paper reduces tthe computational burden for calculating S-transorm using the ideas of sparse FFT. Read more
Published in Applied Acoustics, 2016
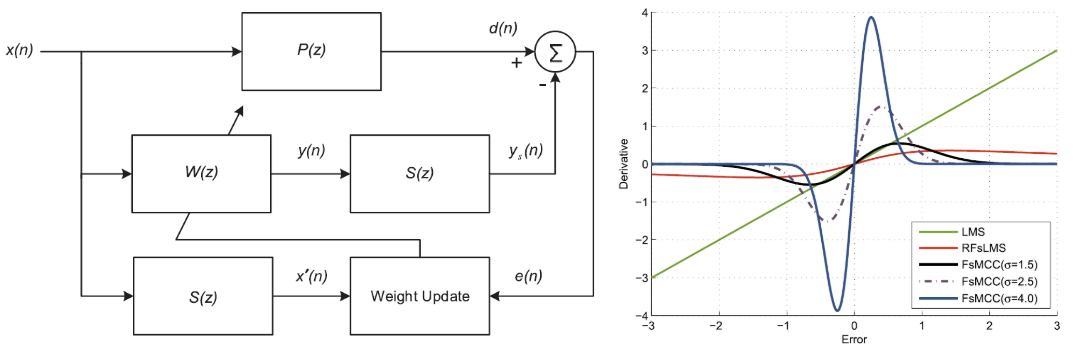
In this paper, we propose a novel information theoretic loss functions for adaptive signal processing under alpha stable distributions. Read more
Published in IEEE TENCON, 2019
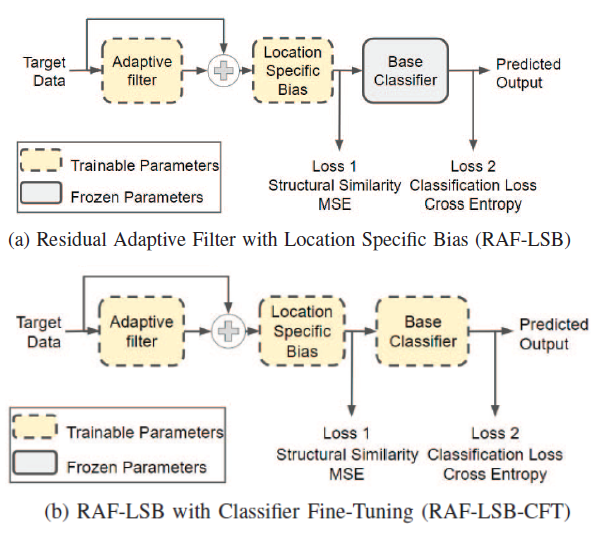
This paper proposes a preprocessing block by adding location specific biases and residual filters that address the generalisation issues between histology datasets. Read more
Published in ESMO MAP, 2019
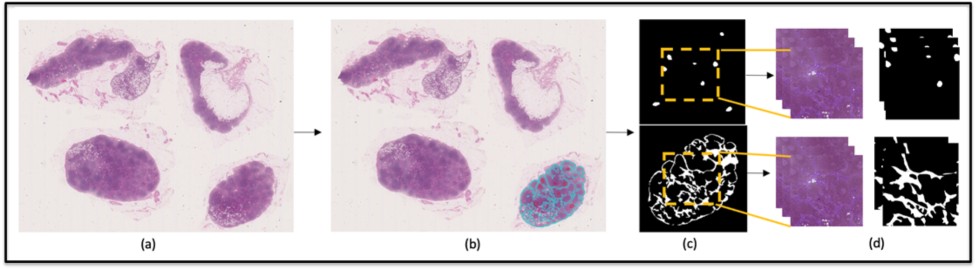
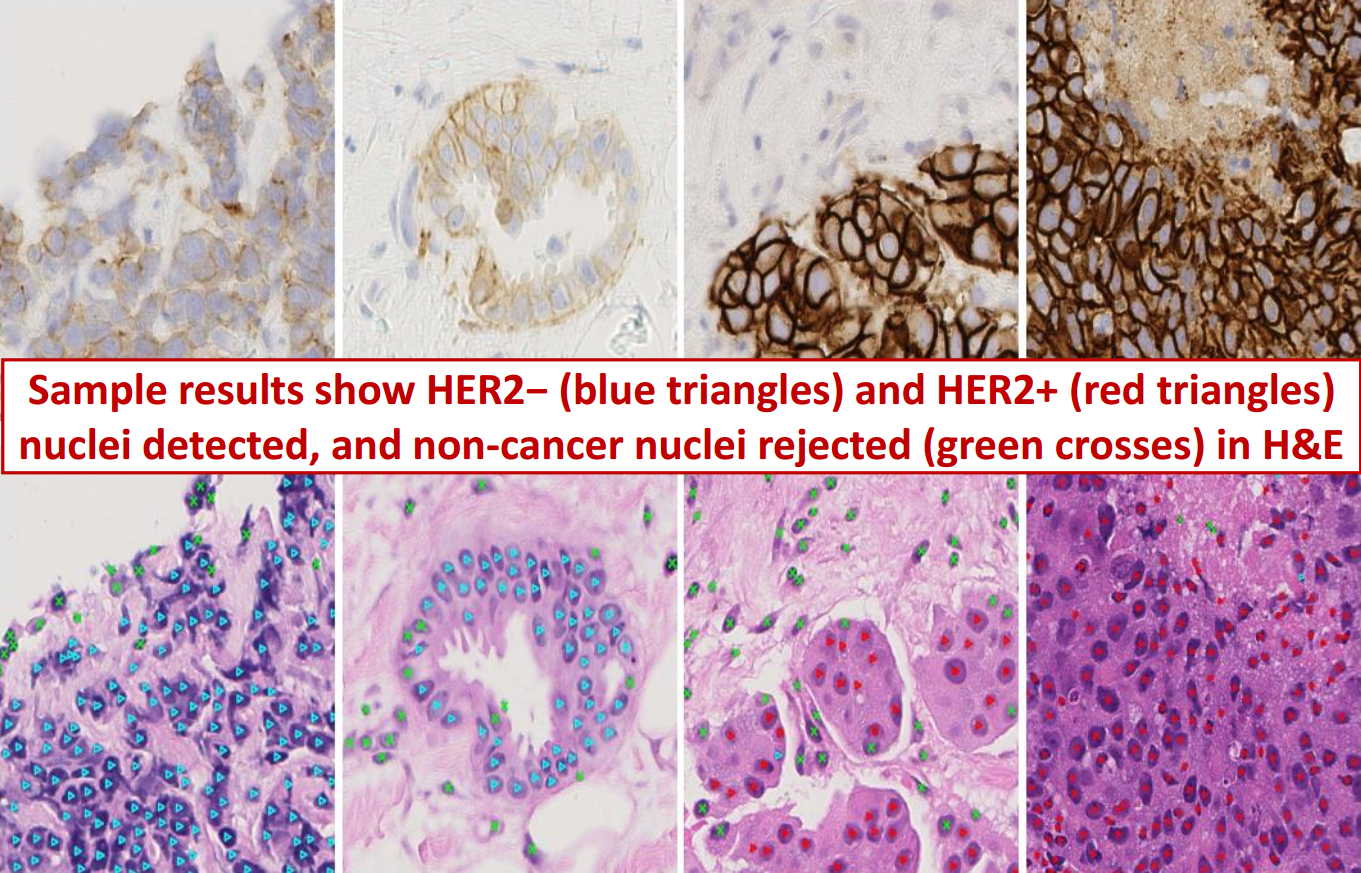
This paper provides a CNN based detection of pathological features in lymph nodes to analyse early signs of breast cancer Read more
Published in Journal of Pathology Informatics, 2019
This paper provides a proof-of-concept for detection of genetic mutation in breast cancer patients from H&E images instead of using costly IHC images. Read more
Published in IGARSS 2020 - 2020 IEEE International Geoscience and Remote Sensing Symposium, 2020

This paper presents a deep learning pipeline for assessing bathymetry maps from satellite derived aerial images . Read more
Published in AACR Virtual Special Conference on Artificial Intelligence, Diagnosis, and Imaging;, 2020
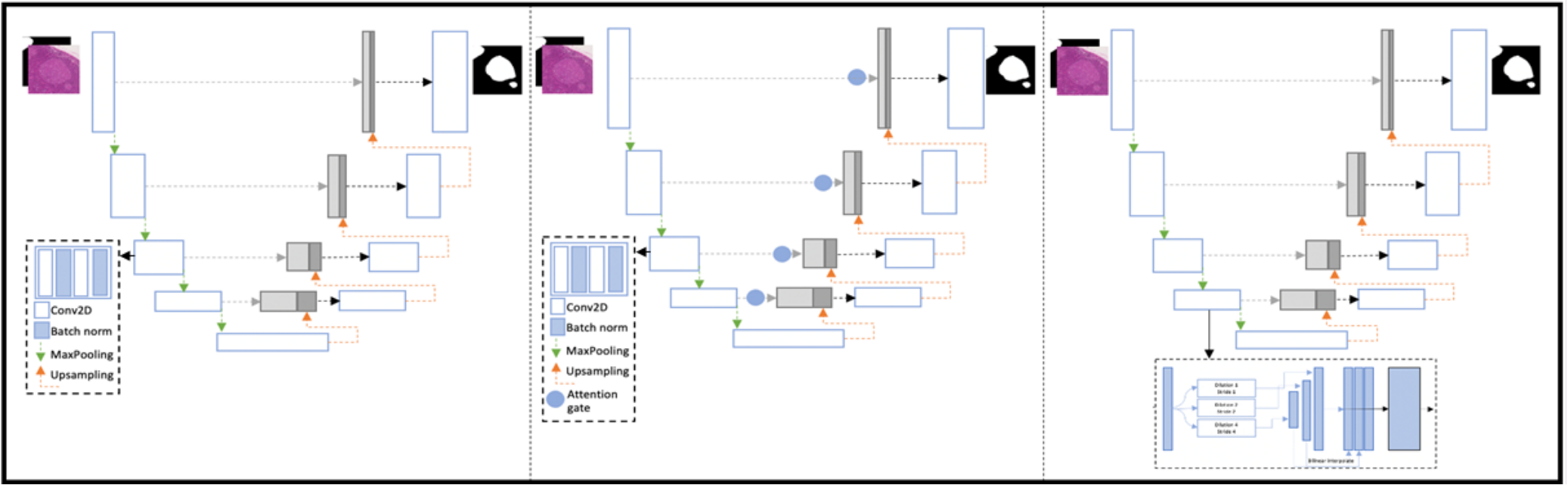
In this work, we propose to classify cancers using graph convolutional networks (GCNs) by modeling a tissue section as a multi-attributed spatial graph of its constituent cells. Read more
Published in Transaction of Medical Imaging, IEEE, 2021
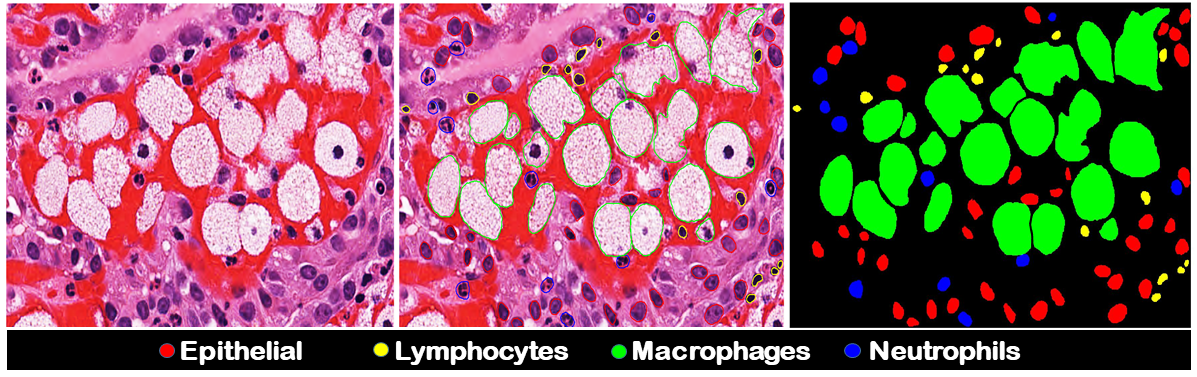
In this paper, we present the key findings of MoNuSAC challenge at ISBI 2020 Read more
Published in Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, 2021
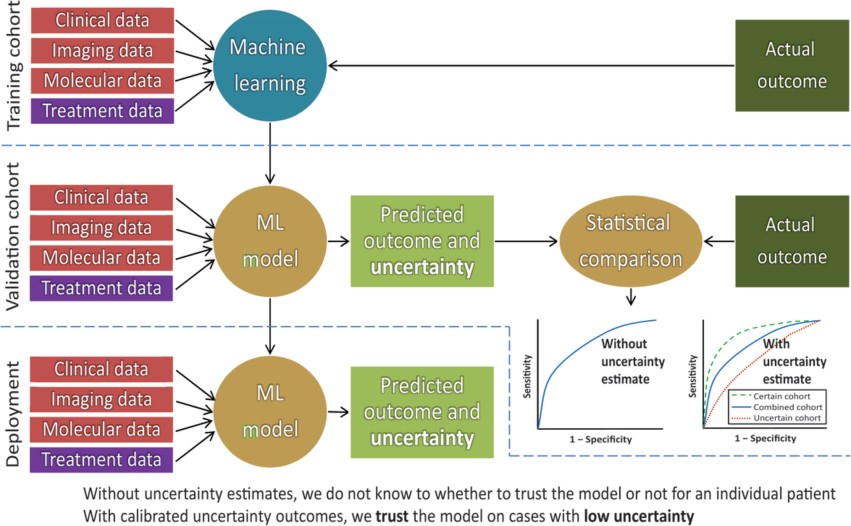
A review on the latest updates on cancer image analytics with deep learning. Read more
Published in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), 2021
This paper introduces novel self-supervision based color-normalisation schemes for H&E images. Read more
Published in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), 2021
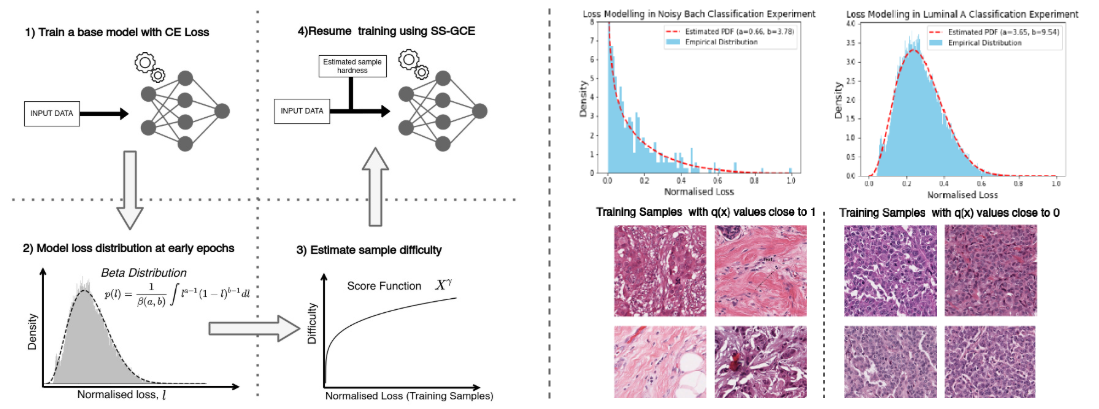
This paper introduces robust sample specific loss functions for histology image analysis. Read more
Published:
Conference proceedings on our paper: “Time frequency analysis: a sparse S transform approach” Abstract:
S transform, which is a powerful time frequency analysis method, has found applications in diverse areas of science and technology. The computational load offered by the S transform increases with increase in the length of the time series which is analysed. In an endeavour to reduce the computational load for time series which is sparse in the frequency domain, a new method for S transform computation is proposed in this paper. The new method uses an efficient search method to identify significant frequency indices and computes the S transform only at the selected frequency indices, thus reducing the computational burden. A simulation study has been carried out to test the efficiency of the proposed method for analytic and real-life signals. The proposed scheme has been shown to provide good signal reconstruction accuracy at a reduced computational load. Read more
Published:
 Conference proceedings on our paper: “Improving Histopathology Classification using Learnable Preprocessing” Abstract:
Conference proceedings on our paper: “Improving Histopathology Classification using Learnable Preprocessing” Abstract:
A deep learning classifier trained on a source dataset often performs poorly on a target dataset, even for the same classification task, due to the differences in the distributions of the two datasets. In histopathology, the problem of dataset bias is even more severe due to the differences in specific tissue preparation and imaging set ups across patient cohorts. With the objective of improving the generalization across datasets, we propose a set of learnable preprocessing operations - an approach that has not been extensively explored - for a supervised deep learning framework that can be trained separately or together with the rest of the neural network. Through preprocessing, the data from a target domain is transformed before being fed to a classification module trained on the source domain to increase the overlap of the former's distribution with that of the latter. Through an extensive set of experiments on histopathology and face datasets, we show the particular and general utility of the proposed preprocessing operations for domain adaptation and compare it to previous approaches. Read more
Published:
As a workshop speaker, introducing the convergence of deep learning for automatic histology image anlysis. Over 100 participants from diverse fields inlcuding medical practioners, doctors, engineering graduates attended the workshop. Read more
Published:
 AICTE Sponsored Faculty Development Program(FDP), on the topic ”The confluence of deep learning in histopathology images”, Virtual Program 2021 Read more
AICTE Sponsored Faculty Development Program(FDP), on the topic ”The confluence of deep learning in histopathology images”, Virtual Program 2021 Read more
Published:
The accuracy of deep learning classifiers trained using the cross entropy loss function suffers even when a fraction of training labels are wrong or input images are uninformative. Training images and labels for computational pathology are often noisy due to the difficulty in signal localization and certain disease classifications being subjective discretization of the underlying continuums of disease conditions. For training classifiers robust to input and label noise, we propose a modified and sample-specific version of generalized cross entropy loss. We take advantage of the bootstrapping properties in deep learning models in order to design loss functions that are aware of the difficulty of classifying individual samples, due to either the label noise or the lack of a strong visual signal. We carry out extensive experiments to validate our approach by comparing against the models trained using other loss functions. The superior performance of our methodology in all the experiments support the requirement of using robust loss functions for histology image classification. Read more
Published:
 Speaker at Nvidia GTC 2021. Delivered a talk on the application of deep learning in the medical imaging and presented a novel loss function for robust histopathology image analysis. Read more
Speaker at Nvidia GTC 2021. Delivered a talk on the application of deep learning in the medical imaging and presented a novel loss function for robust histopathology image analysis. Read more
Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.